A new screening process could dramatically accelerate the identification of nanoparticles suitable for delivering therapeutic RNA into living cells. The technique would allow researchers to screen hundreds of nanoparticles at a time, identifying the organs in which they accumulate – and verifying that they can successfully deliver an RNA cargo into living cells.
Based on work known as “DNA barcoding,” the technique inserts unique snippets of DNA into as many as 150 different nanoparticles for simultaneous testing. The nanoparticles are then injected into animal models and allowed to travel to organs such as the liver, spleen or lungs. Genetic sequencing techniques then identify which DNA-labeled nanoparticles have reached specific organs.
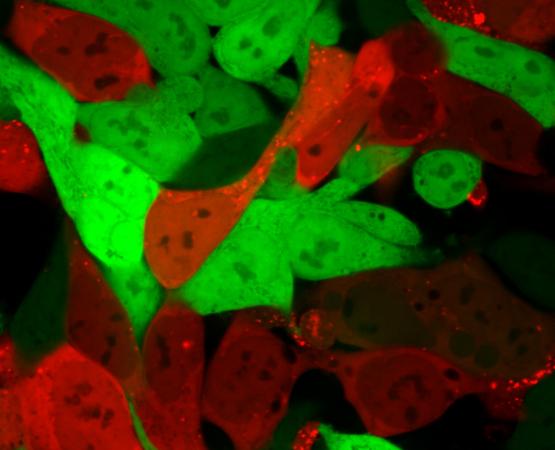
In a paper published October 1 in the journal Proceedings of the National Academy of Sciences, a research team describes taking the process a step farther to verify that the nanoparticles have entered the cells of the specific organs. In addition to the DNA barcode, the researchers inserted into each nanoparticle a snippet of mRNA that is turned into a protein known as “Cre.” The Cre protein generates a red glow, identifying cells that the nanoparticles have entered and successfully delivered the mRNA drug, allowing the researchers to identify which nanoparticles can deliver RNA drugs to the cells of the specific organs.
“This technique, known as Fast Indication of Nanoparticle Discovery (FIND), will allow us to identify the right carrier far more quickly and less expensively than we have been able to do in the past,” said James E. Dahlman, assistant professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. “As a result, the odds that we will be able to find carriers for specific tissues should increase dramatically.”
The FIND technique would replace in vitro screening, which has limited success at identifying nanoparticle carriers for the genetic therapies. The research was supported by funding from the National Institutes of Health, and from the Cystic Fibrosis Research Foundation, the Parkinson’s Disease Foundation and the Bayer Hemophilia Awards Program.
Therapies based on RNA and DNA could address a broad range of genetically based diseases, including atherosclerosis, where such therapies may be able to reverse the buildup of plaque in arteries. Nanoparticles used to deliver RNA and DNA into cells are made from several ingredients whose levels can be varied, creating the potential for tens of thousands of different nanoparticles. Finding the right combination of these ingredients to target specific cells has required extensive trial-and-error discovery processes that have limited the use of RNA and DNA therapies.
Use of the DNA barcoding process allows hundreds of possible nanoparticle combinations to be tested simultaneously in a single animal, but until now, researchers could only tell that the combination had reached specific organs. By examining which cells within the organs have the red glow, they can now verify that the nanoparticles carried the barcodes and delivered functional mRNA drugs into the cells.
In the paper, the researchers report discovering two nanoparticles that efficiently delivered siRNA, sgRNA and mRNA to endothelial cells in the spleen. The researchers believe their technique can deliver therapeutic RNA and DNA to a wide variety of endothelial cell types, and perhaps also to immune system and other cell types.
“The field has been able to functionally deliver genetic drugs to the liver, and we are now trying to use our technology to deliver to different organs and cell types to enable therapies to treat all of the cell types that are in the liver,” said Cory Sago, the paper’s first author and a Ph.D. candidate in Dahlman’s lab. “Now that we have a system that allows us to probe these questions at a very specific level of resolution, we now want to go after other cell types in a more efficient manner.”
Dahlman expects to put the new technology to use quickly.
“We hope to take projects that would ordinarily require years and complete several of them within the next 12 months,” he said. “FIND could be used to carry all sorts of nucleic acid drugs into cells. That could include small RNAs, large RNAs, small DNAs and large DNAs – many different types of genetic drugs that are now being developed in research labs.”
Technical challenges ahead include demonstrating that identifying an affinity for mouse organs predicts which particles will work in the human body, and that the approach works for different classes of genetic therapies.
Experimentally, Dahlman’s lab produces the nanoparticles at three formulation stations that require about 90 seconds to produce each of the 250 or so samples used. The resulting nanoparticles are then examined for proper size range – 40 to 80 nanometers in diameter – before being purified and sterilized for injection into the animals.
After three days, the researchers separate cells that are glowing red and sequence the DNA snippets in them to identify which chemical compositions were most successful at entering cells of specific organs. The most promising chemical compositions are used to develop of a new batch of candidate nanoparticles for a new round of screening, which takes about a week to complete.
“We want to evolve the best particles that we can,” Sago said. “Every single one of the components matters, and we work to get each component right for the cell type that we are interested in. There is a lot of optimization required.”
In addition to those already mentioned, the paper’s co-authors include Melissa P. Lokugamage, Kalina Paunovska, Daryll A. Vanover, Marielena Gamboa Castro, Shannon E. Anderson, Tobi G. Rudoltz, Gwyneth N. Lando, Pooja Tiwari, Jonathan L. Kirschman and Philip J. Santangelo, all of the Coulter Department of Biomedical Engineering; Chris M. Monaco, Young Jang and Nirav N. Shah of the Georgia Tech School of Biological Sciences; Nick Willett of Emory University and the Atlanta Veteran’s Affairs Medical Center, and Anton V. Bryksin of the Parker H. Petit Institute for Bioengineering and Bioscience at Georgia Tech.
The research was supported by the NIH/NIGMS-sponsored Immunoengineering Training Program (T32EB021962), the Georgia Research Assistantship (Grant 3201330), the NIH/NIGMS-sponsored Cell and Tissue Engineering (CTEng) Biotechnology Training Program (T32GM008433), the National Institutes of Health GT BioMAT Training Grant (5T32EB006343), the Cystic Fibrosis Research Foundation (DAHLMA15XX0), the Parkinson’s Disease Foundation (PDF-JFA-1860), and the Bayer Hemophilia Awards Program (AGE DTD). This content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsors.
CITATION: Cory D. Sago, et al., “A high throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing,” (Proceedings of the National Academy of Sciences, 2018) www.pnas.org/cgi/doi/10.1073/pnas.1811276115
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contact: John Toon (404-894-6986) (jtoon@gatech.edu).
Writer: John Toon
Media Contact
John Toon
Research News
(404) 894-6986
Latest BME News
Jo honored for his impact on science and mentorship
The department rises to the top in biomedical engineering programs for undergraduate education.
Commercialization program in Coulter BME announces project teams who will receive support to get their research to market.
Courses in the Wallace H. Coulter Department of Biomedical Engineering are being reformatted to incorporate AI and machine learning so students are prepared for a data-driven biotech sector.
Influenced by her mother's journey in engineering, Sriya Surapaneni hopes to inspire other young women in the field.
Coulter BME Professor Earns Tenure, Eyes Future of Innovation in Health and Medicine
The grant will fund the development of cutting-edge technology that could detect colorectal cancer through a simple breath test
The surgical support device landed Coulter BME its 4th consecutive win for the College of Engineering competition.