One of the biggest questions in science is how life arose from the chemical soup that existed on early Earth. One theory is that RNA, a close relative of DNA, was the first genetic molecule to arise around 4 billion years ago, but in a primitive form that later evolved into the RNA and DNA molecules that we have in life today. New research shows one way this chain of events might have started.
Today, genetic information is stored in DNA. RNA is created from DNA to put that information into action. RNA can direct the creation of proteins and perform other essential functions of life that DNA can’t do. RNA’s versatility is one reason that scientists think this polymer came first, with DNA evolving later as a better way to store genetic information for the long haul. But like DNA, RNA also could be a product of evolution, scientists theorize.
Chemists at the Georgia Institute of Technology have shown how molecules that may have been present on early Earth can self-assemble into structures that could represent a starting point of RNA. The spontaneous formation of RNA building blocks is seen as a crucial step in the origin of life, but one that scientists have struggled with for decades.
“In our study, we demonstrate a reaction that we see as important for the formation of the earliest RNA-like molecules,” said Nicholas Hud, professor of Chemistry and Biochemistry at Georgia Tech, where he’s also the director of the Center for Chemical Evolution.
The study was published Dec. 14 online in the Journal of the American Chemical Society. The research was funded by the National Science Foundation and NASA.
RNA is perfect for the roles it plays in life today, Hud said, but chemically it’s extraordinarily difficult to make. This suggests that RNA evolved from simpler chemical couplings. As life became more chemically complex and enzymes were born, evolutionary pressures would have driven pre-RNA into the more refined modern RNA.
RNA is made of three chemical components: the sugar ribose, the bases and phosphate. A ribose-base-phosphate unit links together with other ribose-base-phosphate units to form an RNA polymer. Figuring out how the bond between the bases and ribose first formed has been a difficult problem to address in the origins of life field, Hud said.
In the study, Hud’s team investigated bases that are chemically related to the bases of modern RNA, but that might be able to spontaneously bond with ribose and assemble with other bases through the same interactions that enable DNA and RNA to store information. They homed in on a molecule called triaminopyrimidine (TAP).
The researchers mixed TAP with ribose under conditions meant to mimic a drying pond on early Earth. TAP and ribose reacted together in high yield, with up to 80 percent of TAP being converted into nucleosides, which is the name for the ribose-base unit of RNA. Previous attempts to form a ribose-base bond with the current RNA bases in similar reactions had either failed or produced nucleosides in very low yields.
“This study is important in showing a feasible step for how we get the start of an RNA-like molecule, but also how the building blocks of the first RNA-like polymers could have found each other and self-assembled in what would have been a very complex mixture of chemicals,” Hud said.
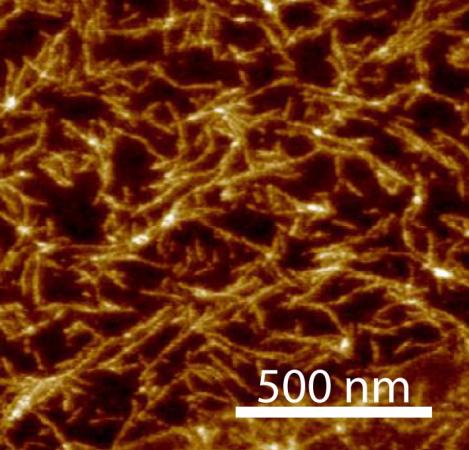
The researchers demonstrated this property of the TAP nucleosides by adding another molecule to their reaction mixture, called cyanuric acid, which is known to interact with TAP. Even in the unpurified reaction mixture, noncovalent polymers formed with thousands of paired nucleosides.
“It is amazing that these nucleosides and bases actually assemble on their own, as life today requires complex enzymes to bring together RNA building blocks and to spatially order them prior to polymerization,”said Brian Cafferty, a graduate student at Georgia Tech and co-author of the study
The study demonstrated one possible way that the building blocks for an ancestor of RNA could have come together on early Earth. TAP is an intriguing candidate for one of the first bases that eventually led to modern RNA molecules, but there are certainly others, Hud said.
Future work, in Hud’s lab and by other laboratories in the Center for Chemical Evolution, will investigate the origins of RNA’s phosphate backbone, as well as other pathways toward modern RNA.
“We’re looking for a simple, robust chemistry that can explain the earliest origin of RNA or its ancestor,” Hud said.
This research is supported by the National Science Foundation (NSF) Center for Chemical Evolution under award number CHE-1004570, and the NASA Exobiology Program under award number NNX13AIO2G. Any conclusions or opinions are those of the authors and do not necessarily represent the official views of the sponsoring agencies.
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contacts: Brett Israel (404-385-1933) (brett.israel@comm.gatech.edu) or John Toon (404-894-6986) (jtoon@gatech.edu)
Writer: Brett Israel
Media Contact
Brett Israel
404-385-1933
Keywords
Latest BME News
Jo honored for his impact on science and mentorship
The department rises to the top in biomedical engineering programs for undergraduate education.
Commercialization program in Coulter BME announces project teams who will receive support to get their research to market.
Courses in the Wallace H. Coulter Department of Biomedical Engineering are being reformatted to incorporate AI and machine learning so students are prepared for a data-driven biotech sector.
Influenced by her mother's journey in engineering, Sriya Surapaneni hopes to inspire other young women in the field.
Coulter BME Professor Earns Tenure, Eyes Future of Innovation in Health and Medicine
The grant will fund the development of cutting-edge technology that could detect colorectal cancer through a simple breath test
The surgical support device landed Coulter BME its 4th consecutive win for the College of Engineering competition.