By the time they’re two, most children have had respiratory syncytial virus (RSV) and suffered symptoms no worse than a bad cold. But for some children, especially premature babies and those with underlying health conditions, RSV can lead to pneumonia and bronchitis – which can require hospitalization and have long-term consequences.
A new technique for studying the structure of the RSV virion and the activity of RSV in living cells could help researchers unlock the secrets of the virus, including how it enters cells, how it replicates, how many genomes it inserts into its hosts – and perhaps why certain lung cells escape the infection relatively unscathed. That could provide scientists information they need to develop new antiviral drugs and perhaps even a vaccine to prevent severe RSV infections.
“We want to develop tools that would allow us to get at how the virus really works,” said Philip Santangelo, an associate professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. “We really need to be able to follow the infection in a single living cell without affecting how the virus infects its hosts, and this technology should allow us to do that.”
The research was supported by the National Institutes of Health’s National Institute of General Medical Sciences and published online ahead of print in the journal ACS Nano on December 30, 2013. While RSV will be the first target for the work, the researchers believe the imaging technique they developed could be used to study other RNA viruses, including influenza and Ebola.
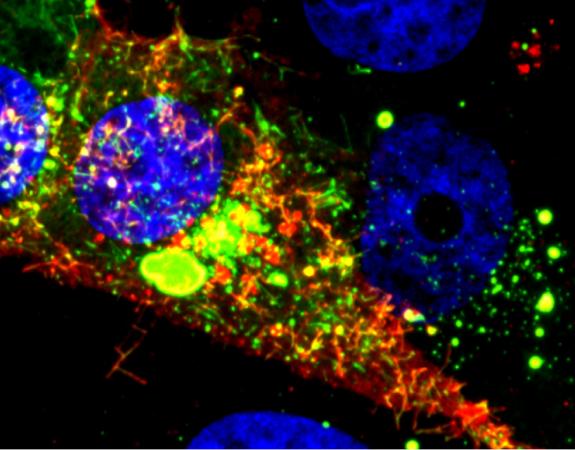
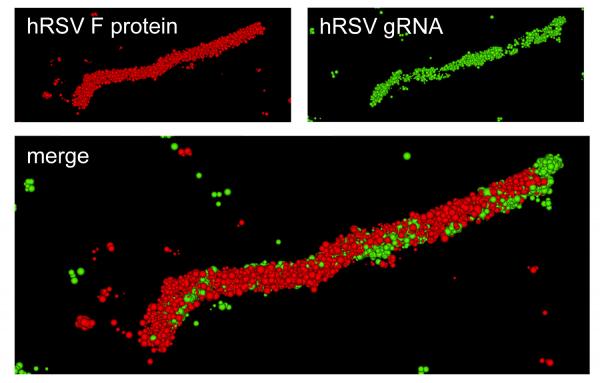
“We’ve shown that we can tag the genome using our probes,” explained Santangelo. “What we’ve learned from this is that the genome does get incorporated into the virion, and that the virus particles created are infectious. We were able to characterize some aspects of the virus particle itself at super-resolution, down to 20 nanometers, using direct stochastic optical reconstruction microscopy (dSTORM) imaging.”
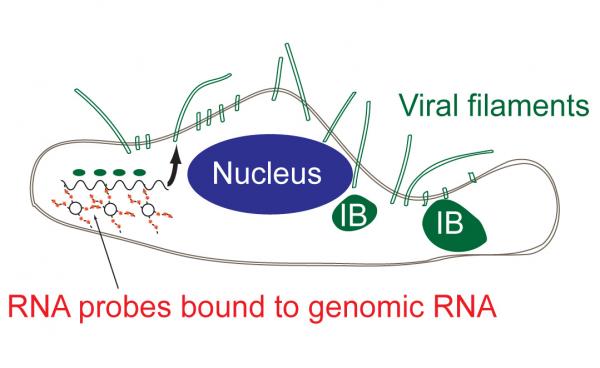
RSV can be difficult to study. For one thing, the infectious particle can take different forms, ranging from 10-micron filaments to ordinary spheres. The virus can insert more than one genome into the host cells and the RNA orientation and structure are disordered, which makes it difficult to characterize.
The research team, which included scientists from Vanderbilt University and Emory University, used a probe technology that quickly attaches to RNA within cells. The probe uses multiple fluorophores to indicate the presence of the viral RNA, allowing the researchers to see where it goes in host cells – and to watch as infectious particles leave the cells to spread the infection.
“Being able to see the genome and the progeny RNA that comes from the genome with the probes we use really give us much more insight into the replication cycle,” Santangelo said. “This gives us much more information about what the virus is really doing. If we can visualize the entry, assembly and replication of the virus, that would allow us to decide what to go after to fight the virus.”
The research depended on a new method for labelling RNA viruses using multiply-labeled tetravalent RNA imaging probes (MTRIPS). The probes consist of a chimeric combination of DNA and RNA oligonucleotide labeled internally with fluorophores tetravelently complexed to neutravidin. The chimeric combination was used to help the probes evade cellular defenses.
“There are lots of sensors in the cell that look for foreign RNA and foreign DNA, but to the cell, this probe doesn’t look like anything,” Santangelo explained. “The cell doesn’t see the nucleic acid as foreign.”
Introduced into cells, the probes quickly diffuse through a cell infected with RSV and bind to the virus’s RNA. Though binding tightly, the probe doesn’t affect the normal activities of the virus and allows researchers to follow the activity for days using standard microscopy techniques. The MTRIPS can be used to complement other probe technology, such as GFP and gold nanoparticles.
Work done by graduate student Eric Alonas to concentrate the virus was essential to the project, Santangelo said. The concentration had to be done without adversely affecting the infectivity of the virus, which would have impacted its ability to enter host cells.
“It took quite a bit of work to get the right techniques to concentrate the RSV,” he said. “Now we can make lots of infectious virus that’s labelled and can be stored so we can use it when we want to.”
To study the infection’s progress in individual cells, the researchers faced another challenge: living cells move around, and following them complicates the research. To address that movement, the laboratory of Thomas Barker – also in the Coulter Department – used micro-patterned fibronectin on glass to create 50-micron “islands” that contained the cells during the study.
Among the mysteries that the researchers would like to tackle is why certain lung cells are severely infected – while others appear to escape ill effects.
“If you look at a field of cells, you see huge differences from cell to cell, and that is something that’s not understood at all,” Santangelo said. “If we can figure out why some cells are exploding with virus while others are not, perhaps we can figure out a way to help the bad ones look more like the good ones.”
In addition to those already mentioned, the research team included James Crowe, professor of pediatrics at Vanderbilt University; Elizabeth Wright, assistant professor in the School of Medicine at Emory University; Daryll Vanover, Jeenah Jung, Chiara Zurla, Jonathan Kirschman, Vincent Fiore, and Alison Douglas from the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University; Aaron Lifland and Manasa Gudheti from Vutara Inc. in Salt Lake City, and Hong Yi from the Emory University School of Medicine.
One of the challenges of studying RSV is maintaining its activity in the laboratory setting – a problem parents of young children don’t share.
“When you handle this virus in the lab, you have to always be careful about it losing infectivity,” Santangelo noted. “But if you take a room full of children who have not been infected and let one infected child into the room, 15 minutes later all of the children will be infected.”
The research described here was supported by the National Institute of General Medical Sciences of the National Institutes of Health under contract R01 GM094198-01. Any conclusions or opinions expressed are those of the authors and do not necessarily represent the official views of the NIH.
CITATION: Eric Alonas, et al., “Combining Single RNA Sensitive Probes with Subdiffraction-Limited and Live-Cell Imaging Enables the Characterization of Virus Dynamics in Cells,” (ACS Nano, December 2013). (http://dx.doi.org/10.1021/nn405998v).
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contacts: John Toon (jtoon@gatech.edu) (404-894-6986) or Brett Israel (brett.israel@comm.gatech.edu) (404-385-1933).
Writer: John Toon
Media Contact
John Toon
Research News
(404) 894-6986
Keywords
Latest BME News
Jo honored for his impact on science and mentorship
The department rises to the top in biomedical engineering programs for undergraduate education.
Commercialization program in Coulter BME announces project teams who will receive support to get their research to market.
Courses in the Wallace H. Coulter Department of Biomedical Engineering are being reformatted to incorporate AI and machine learning so students are prepared for a data-driven biotech sector.
Influenced by her mother's journey in engineering, Sriya Surapaneni hopes to inspire other young women in the field.
Coulter BME Professor Earns Tenure, Eyes Future of Innovation in Health and Medicine
The grant will fund the development of cutting-edge technology that could detect colorectal cancer through a simple breath test
The surgical support device landed Coulter BME its 4th consecutive win for the College of Engineering competition.