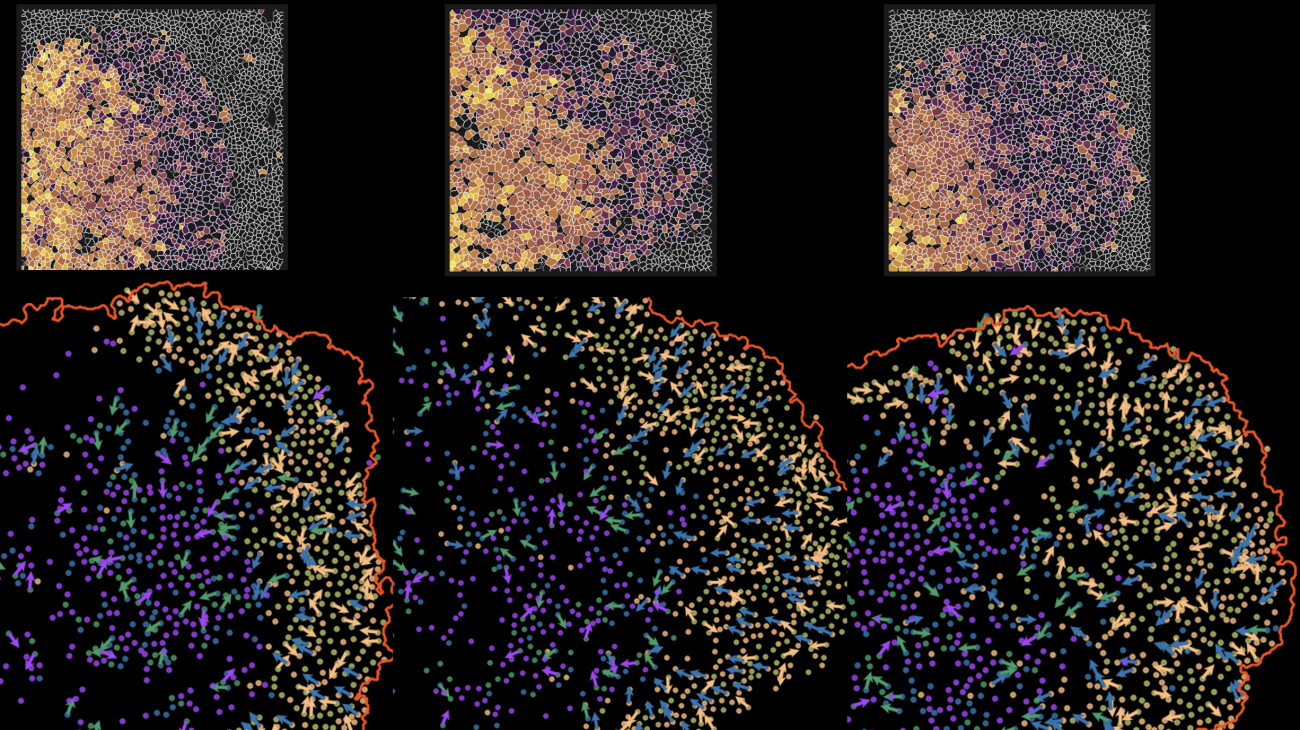
Images of time in space: The top panel image shows pseudo-time single cell metabolic trajectories across distinct biogeographical regions. The dark purple represents early metabolic changes, while the bright yellow represents later metabolic activities. The bottom panel is a spatial projection of single cells’ metabolic trajectories (denoted by arrows in the dark zone and light zone regions) in tonsil tissue. Photo provided by Coskun Lab
By Jerry Grillo
Ahmet Coskun and his collaborators plan to create a chemical atlas of all the immune cells in the human body, a 3D micromap to help clinicians navigate the complex role of the entire immune system in the presence of different diseases.
It’s the kind of massive undertaking that would result in vastly improved precision therapies for patients. And it’s the kind of journey that starts with a single cell. Coskun and team are off to a fast start with the introduction of a new integrative technique for profiling human tissue that enables researchers to capture the geography, structure, movement, and function of molecules in a 3D picture.
The researchers described their new approach, the Single Cell Spatially resolved Metabolic (scSpaMet) framework, in the journal Nature Communications on Dec. 13. The study builds on a technique Coskun’s team developed and described in a 2021 article, “3D Spatially resolved Metabolomic profiling Framework,” published in Science Advances. In that work, the team introduced a technique that measures the activity of metabolites and proteins as part of a comprehensive profile of human tissue samples.
“Earlier we couldn’t achieve single-cell resolution, but with this new approach, we can,” said Coskun, Bernie Marcus Early Career Professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. “With this new approach, we can get spatial details of proteins and metabolites in single cells– no one else has yet reached this level of high subcellular resolution.”
He added, “We’re pioneering a new field of research with this work, single cell spatial metabolomics.”
A Bigger, Better Molecular Picture
Human tissue is spatially crowded with all kinds of stuff, so investigators need tools that can see clearly into, through, and around that multilayered biological traffic – everything, all at once, in high-definition 3D. With scSpaMet, Coskun’s team can capture single cell details such as the naturally occurring lipids, proteins, as well as metabolites (with their multiple functions, including energy conversion and cell signaling). And other details, like those provided by researchers: Intracellular and surface markers are used to label and track cell activity and behavior.
The team broadened the scope of this study, extending its investigation beyond human tonsil tissue.
“We showed the crucial role of immune cells in lung cancer for the study of lung cancer for the study of immunometabolism of T cells and macrophages as they interact with tumors,” Coskun said. “Then we created dynamic immune metabolic changes in tonsils as they go through germinal center reactions to give rise to the antibody-producing cells. Finally, we demonstrated the role of immune cells in the endometrium, a membrane in the uterus that might lead to conditions impacting a woman’s health.”
The wide-angled study required plenty of cross-country collaboration with other institutions, although Coskun’s lab guided the wide-angled study, integrating its expertise in bioimaging, chemistry, tissue biology, and artificial intelligence.
Cold Spring Harbor Laboratory (New York) provided access to its endometrium tissue bank. Oak Ridge National Laboratory (Tennessee) provided data from its complex metabolic imaging instrumentation, to further demonstrate how single cell spatial metabolomics imaging can generate rich data.
The University of California-Davis provided kidney biospecimens as both fixed tissue and frozen embedded tissue, in two halves of the same sample, “so we could demonstrate the effect of tissue preparation on the sensitivity of our single cell spatial metabolomics pipeline,” Coskun said.
The team also included Thomas Hu and Mayar Allam, graduate researchers in Coskun’s lab, who guided the research as lead authors, and Walter Henderson, a research scientist who manages the IEN/IMat Materials Characterization Facility at Georgia Tech.
Considering the Whole Person's Biochemistry
The ability to generate single cell spatial metabolic profiling of individual patients can reveal a world of possibility and potential for clinicians who need to fully understand a patient’s biophysical makeup to contrive the best treatment options.
“For example, it can provide mechanisms of how immune responses can be boosted by adding dietary molecules along with immunotherapies,” Coskun said. “It can also help adjust the dose of cell-based treatments, based on the body mass index of individual patients, whether they are obese or not.”
Coskun believes this new arena of single cell metabolomics research his lab is developing will complement the field of single cell genomics, which has led to genomic medicine. His team’s comprehensive exploration and imaging of the geography of normal and unhealthy human tissues – of every single cell – can further explain cellular regulation in ways that were previously overlooked, due to the lack of technology.
He envisions a future in which a patient’s BMI, dietary habits, and exercise commitments, along with their single cell spatial metabolomic atlas of disease progression, will be analyzed all together to find optimum therapies that can work with biologics and metabolic boosting regimens, potentially increasing the survival of cancers, women’s diseases, and metabolic disorders.
“We will have opportunities to talk about spatial single cell metabolomic medicine, to stratify patients and design next-generation combination therapies with an integrated view of genes and chemical activity roadmaps, for more efficient management of cancer and other diseases,” Coskun said.
In creating their scSpaMet framework, the researchers must integrate expensive machines that live in the worlds of nanotechnology and chemistry right now. The system will require clinical-friendly optimizations to be able to run single cell metabolic imaging measurements in healthcare settings. Coskun expects the cost and user-friendliness will be improved in the near future to reach the bedside.
“When researchers achieved single cell sequencing, it was a revolutionary moment in medicine,” Coskun said. “Now, we believe single cell spatial metabolic profiling will push the medical practice into new heights.”
This research was supported by the Burroughs Wellcome Fund, and the Bernie Marcus Early Career Professorship, as well as the National Science Foundation (Grant ECCS-1542174), (Grant ECCS-2-25462), American Cancer Society, and National Institutes of Health grants (R21AG081715, R21AI173900, and R35GM151028)
Citation: Thomas Hu, Mayar Allam, Shuangyi Cai, Walter Henderson, Brian Yueh, Aybuke Garipcan, Anton V. Ievlev, Maryam Afkarian, Semir Beyaz, and Ahmet F. Coskun. “Single-cell spatial metabolomics with cell-type specific protein profiling for tissue systems biology,” Nature Communications (Dec. 13, 2023)
Latest BME News
Jo honored for his impact on science and mentorship
The department rises to the top in biomedical engineering programs for undergraduate education.
Commercialization program in Coulter BME announces project teams who will receive support to get their research to market.
Courses in the Wallace H. Coulter Department of Biomedical Engineering are being reformatted to incorporate AI and machine learning so students are prepared for a data-driven biotech sector.
Influenced by her mother's journey in engineering, Sriya Surapaneni hopes to inspire other young women in the field.
Coulter BME Professor Earns Tenure, Eyes Future of Innovation in Health and Medicine
The grant will fund the development of cutting-edge technology that could detect colorectal cancer through a simple breath test
The surgical support device landed Coulter BME its 4th consecutive win for the College of Engineering competition.